Microbial Genomics: Standing on the Shoulders of Giants
The Institute for Genomic Research (TIGR)

TIGR’s bioinformatics team pioneered some of the earliest programs for genome assembly and annotation in microbial genomics. Researchers who transited through TIGR early in their careers say it was a life-changing experience, and to this day feel a kinship that borders on cult. Four TIGR alumni – Elodie Ghedin and Jane Carlton (New York University), Neil Hall (Earlham Institute), Adam Phillippy (National Human Genome Research Institute/NIH) – reminisce on their time at this extraordinary institute.
Riding the TIGR – an oral history
The Institute for Genomic Research (TIGR, pronounced ‘tiger’) was a nonprofit, private research institute founded in 1992 by J. Craig Venter and Claire Fraser. In 2006, it merged with the J. Craig Venter Institute and disappeared into the pages of history. Although its lifespan was shorter than that of the average tiger, it made a huge impact on the field of genomics. Venter and Fraser were both researchers at the National Institutes of Health (NIH) when, seeking more flexibility and access to newer technology than the NIH was willing to provide, they created TIGR. In 1993, the first full year of operation, TIGR sequenced over 101,000 samples, and with twenty ABI 373 sequencers it was considered the largest sequencing facility in the world. Shotgun sequencing was first developed at TIGR and used to decode the whole genome of a bacterium (Haemophilus influenza), costing approximately $750,000 over 13 months. In fact, TIGR has a long list of firsts in the microbial sequencing field (Fig. 1): the first bacterial genome, the first archaeal genome, the first parasite chromosome, the first large-scale viral genomes project, the first parasitic nematode genome, and many other microbial pathogens. TIGR’s bioinformatics team pioneered some of the earliest programs for genome assembly and annotation in microbial genomics, such as TIGR Assembler, GLIMMER, and MUMmer. Researchers who transited through TIGR early in their careers say it was a life-changing experience, and to this day feel a kinship that borders on cult. Four TIGR alumni – Elodie Ghedin and Jane Carlton, now at New York University; Neil Hall at the Earlham Institute in the UK; and Adam Phillippy at the National Human Genome Research Institute/NIH – reminisce on their time at this extraordinary institute.
Elodie Ghedin: When you Google ‘TIGR’, the first thing that pops up is some type of bike lock device. And when you tell millennials that you once worked at TIGR, you are met with a blank stare, and a ‘what’s TIGR?’. That, more than having a child now in her twenties, has made me feel like my heydays are in the past because I remember when saying you worked at TIGR led to being simultaneously respected and despised. I joined TIGR in 2000 as a postdoc working on the genomes of the parasites Trypanosoma brucei and Trypanosoma cruzi. TIGR was part of the TriTryp genome conglomerate (the third being Leishmania major) with the Sanger Institute (now called the Wellcome Trust Sanger Institute), the Seattle Biomedical Research Institute (now called the Center for Infectious Disease Research), and the Karolinska Institute. Najib El-Sayed, now at University of Maryland, led the TIGR effort. Jane, didn’t you join a year later?
Jane Carlton: That's right, I joined the TIGR faculty in 2001 from NCBI (the National Center for Biotechnology Information at NIH), just as the projects for the first malaria parasite genomes took shape. This was such an exciting time! The sheer exuberance at uncovering the biology of an organism for the very first time was palpable. In the Parasite Group, we churned out the first reference genomes for several of the major eukaryotic microbes that cause devastating human and animal diseases, identifying possible new drug and vaccine molecules along the way – it was ground-breaking.
Adam Phillippy: Jane was one of my first research mentors! It was a formative experience for me. I was a summer intern for Steven Salzberg (now at Johns Hopkins) in 2001, who was TIGR’s director of bioinformatics at the time. Steven wanted me to work with Jane on aligning different Plasmodium genomes to one another, but there weren’t a lot of tools available. So, I spent the summer hacking MUMmer in a ‘portable’ office set up in the parking lot because there was no room left in the buildings. I wrote the first version of the Nucmer/Promer alignment suite that summer, jamming out to Sasha and Digweed. I still remember pouring over *printed* alignment output with Jane and Steven. I was just a naive kid from Pennsylvania, and here was the esteemed Dr. Carlton calling my results ‘brilliant’! (Jane was my first British collaborator, so it took me a while to figure out that she wasn’t that impressed.) I already knew I wanted to do genomics, but that summer completely hooked me. The next year I turned down acceptance to a PhD programme and joined Steven’s team full time. It was an absolute dream job – füssball every lunch break and keggers at the campus cafeteria on Fridays. I could have lived that life forever. Everyone I worked with turned into a lifelong friend, and I’m one of the many alums that married a fellow TIGR and had some TIGR cubs (as Claire Fraser once called them). I expect great things from that generation of kids!
Neil Hall: I moved from the Sanger Institute to TIGR in 2003. This was quite a bold move at the time as there was a huge rivalry between the institutes, particularly as NIH and the Wellcome Trust forced collaboration upon us by jointly funding pathogen genome projects. However, during these collaborations many TIGR faculty became my best frenemies and so I made the jump to the TIGR bioinformatics group. This was led by Owen White, now at the Institute for Genome Sciences (IGS) at the University of Maryland, and Steven Salzberg. These guys made the most unlikely team: Owen was (is) a laid-back drummer, artist and charismatic new-age philosopher who proudly described himself as a ‘blue collar bioinformatician’. Steven was (is)… well… none of those things. But around them they formed a nucleus of brilliant scientists covering algorithm development through to annotation and data curation. There was a huge group supporting a few faculty PIs (Principal Investigators), possibly over 80 bioinformaticians. But it worked because of a shared respect for the data and a pride in generating ‘the best’ microbial genomes. Or maybe it worked because of a shared passion for füssball (which is table football in old English) and multiplayer gaming (mostly ‘Doom’) after 3pm. Or maybe our motivation came from the chainsaw on the wall of Owen’s office with the words ‘Management tool’ inscribed on it…
Adam: Most of my bioinformatics work was to support comparative genomics for the ‘massive’ number (>10) of microbial genomes TIGR was doing at the time. My first assignment after joining Steven’s group was to work with Mihai Pop (University of Maryland), developing bioinformatics tools for the FBI's ‘Amerithrax’ investigation, which sought the source of the 2001 anthrax mailings. TIGR’s role was to sequence and compare a bunch of Bacillus anthracis genomes collected from the investigation. Conveniently, I had just written whole-genome alignment tools for Jane’s project that summer, and so we ported those tools from malaria to anthrax. It was a bit surreal, because we were interfacing with Jacques Ravel (now at IGS), who was sworn to secrecy by the FBI and couldn’t tell us what samples we were analysing. So, we’d report some result for an anonymised B. anthracis genome and he’d get all excited and go running off and we’d have no idea why. Looking back, TIGR was working on the first ever genomic reconstruction of an infectious disease outbreak, and I didn’t even know it. I was just in the trenches writing code (and playing füssball).
Neil: As I remember it Adam was scarily good at füssball – he is possibly wasted in computational biology!
Elodie: One of my favorite parts was the insertion of hidden icons in the TIGR genome annotation figures. The first was ‘Elvis Lives’ hidden in the 1995 Haemophilus annotation (Fig. 2a). However, having been tipped, in subsequent submissions the editors were good at spotting the icons after squinting at 400X blow-ups of figures. That’s how they found the little ‘etch-a-sketch’ Bart Vader (Fig. 2b) (for a collaborator at the Sanger Institute) in the P. falciparum genome paper [11]. The comment was ‘we found an image in the figure that was not referred to in the legend, so we removed it’. Najib (El-Sayed) tried to hide a tiny chicken in the T. brucei genome figure published in Science in 2005. This was an inside joke to poke fun at Nature editors who had wanted to gut our paper by restricting it to only 2000 words when they had just published a ‘tome’ on the chicken genome (the chicken!!). We had turned to Science instead, but were still miffed.
Jane: Yes, that was our form of #Resistance! Malcolm Gardner (University of Washington School of Public Health) and Hervé Tettelin (IGS) tried to insert a mosquito icon in the Plasmodium falciparum chromosome 2 paper in 1998 but an editor spotted it. We also tried to insert a human/monkey glyph for the Plasmodium vivax genome paper for a collaborator who was ‘monkeying’ us around. We almost got one for the sexually transmitted parasite Trichomonas vaginalis (guess what the icon was for that!) but in the end the genome assembly was too fragmented for a figure.
Neil: No, no, the mosquito did make it in. I haven't found it BUT I SWEAR there is a mosquito in the P. falciparum chromosome 2 paper. And did you all know that E. A. Presley worked at TIGR? https://www.ncbi.nlm.nih.gov/nuccore/NC_001733.1
Adam: It's hard to pin down exactly why TIGR was so special, but funding was plentiful and we were breaking new ground every day. It had a successful start-up feel. Funding is so much tighter these days, so it’s hard to recreate that kind of creativity and playfulness. In the end, it’s the people that I remember the most. A concentrated group of bright, energetic, diverse people all working on something special. Science at its best.
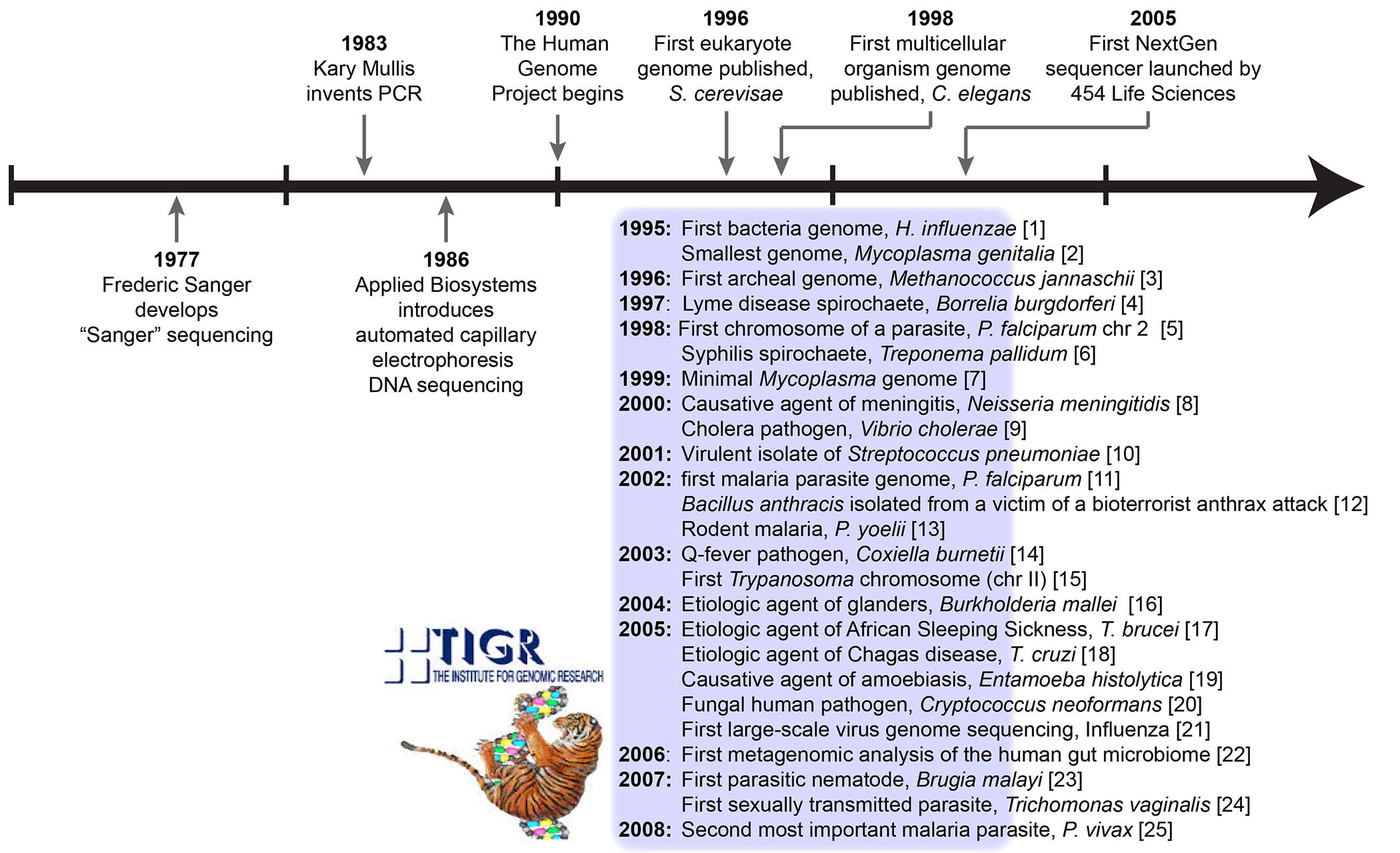
Figure 1. Timeline of events in the history of genomics with some of TIGR's contributions, primarily in the field of microbial pathogens.
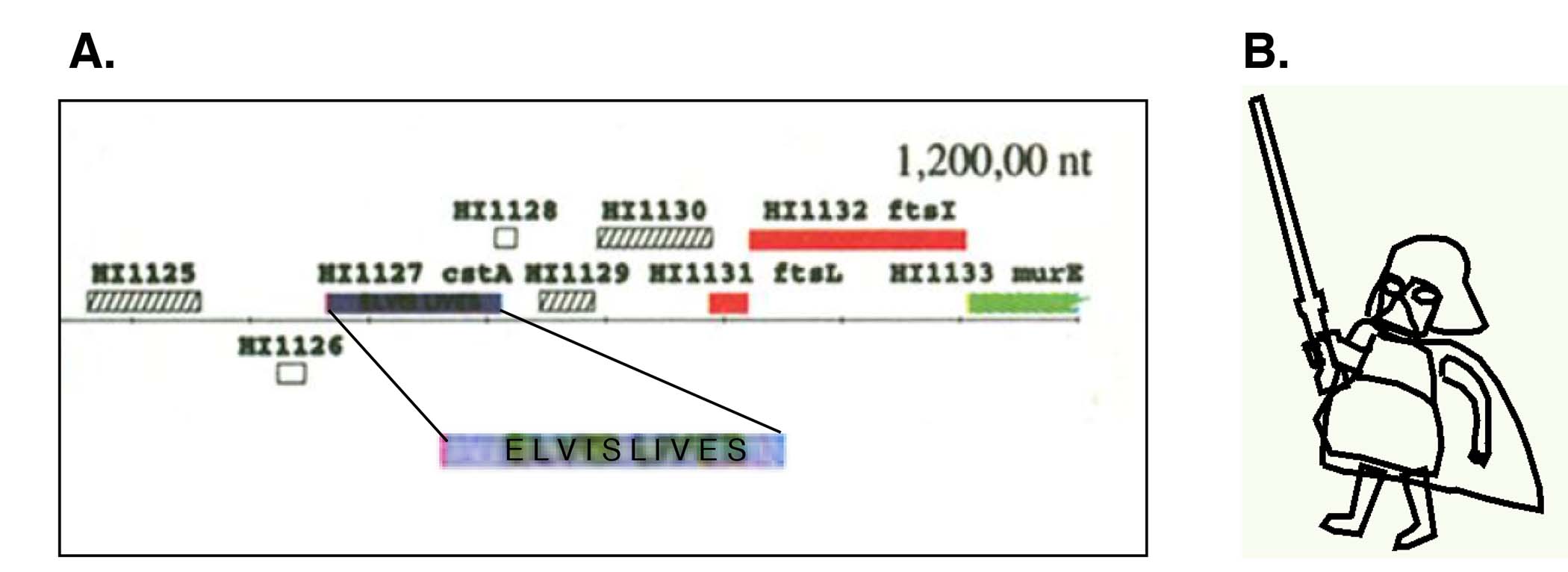
Figure 2. (a) ‘Message’ hidden in the annotation figure for the first bacterial genome published [1]; (b) icon that was removed by Nature editors from the figure of the P. falciparum genome [11].
References
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 1995;269:496–512
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA et al. The minimal gene complement of Mycoplasma genitalium. Science 1995;270:397–403
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 1996;273:1058–1073
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 1997;390:580–586
- Gardner MJ, Tettelin H, Carucci DJ, Cummings LM, Aravind L et al. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science 1998;282:1126–1132
- Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 1998;281:375–388
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 1999;286:2165-2169
- Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58 Science 2000;287:1809–1815
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 2000;406:477–83
- Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 2001;293:498–506
- Gardner MJ, Hall N, Fung E, White O, Berriman M et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002;419:498–511
- Read TD, Salzberg SL, Pop M, Shumway M, Umayam L et al. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 2002;296:2028–2033
- Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 2002;419:512–509
- Seshadri R, Paulsen IT, Eisen JA, Read TD, Nelson KE et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci USA 2003;100:5455–5460
- El-Sayed NM, Ghedin E, Song J, MacLeod A, Bringaud F et al. The sequence and analysis of Trypanosoma brucei chromosome II. Nucleic Acids Res 2003;31:4856-4863
- Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE et al. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci USA. 2004;101:14246–14251
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H et al. The genome of the African trypanosome Trypanosoma brucei. Science 2005;309:416–422
- El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 2005;309:409–415
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J et al. The genome of the protist parasite Entamoeba histolytica. Nature 2005;433:865–868
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 2005;307:1321–1324
- Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 2005;437:1162–1166
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ et al. Metagenomic analysis of the human distal gut microbiome. Science 2006;312:1355–1359
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q et al. Draft genome of the filarial nematode parasite Brugia malayi. Science 2007;317:1756–1760
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 2007;315:207–212
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 2008;455:757–763
To read previous SOTSOG pieces, click here.
